Writer Control#
Overview#
Writers determines what data to output and its format. Isaacsim.Replicator.Agent (IRA) provides built-in writers as well as the ability for you to create your own custom writers.
All the IRA built-in writers are derived from IRABasicWriter, which is derived from the Replicator BasicWriter. As a result, all the annotators from the Replicator BasicWriter are supported with the following exceptions:
bounding_box_2d_tight,bounding_box_2d_lose,bounding_box_3dare overwritten by annotators with corresponding names starting with object_info_.skeleton_datais overwritten byagent_info_skeleton_data.S3 related parameters are disabled for now.
In addition, while data written is identical from Replicator, IRA built-in writers output each annotator data to separated folders for better readibility. The annotators overwritten by the IRABasic writer are organized into one file called object_detection.json.
Built-in Writers#
IRABasicWriter#
Overview#
IRABasicWriter is the base writer for all IRA built-in writers and it is derived from the Replicator BasicWriter.
Parameters#
By default, all the following annotators are turned off. The output data is written to the object_detection.json file.
object_info_bounding_box_2d_tight: outputs objects bounding box 2d tight data. The output data is written to the object section.object_info_bounding_box_2d_lose: outputs objects bounding box 2d loose data. The output data is written to the object section.object_info_bounding_box_3d: outputs objects bounding box 3d data. The output data is written to the object section.agent_info_skeleton_data: outputs agents skeleton data. The output data is written to agent section.More sections in the UI display the parameters inherited from BasicWriter.
rgbandcamera_paramsare on by default while other annotators follow the Replicator’s default setting. Refer to Replicator Annotators Information for details.
Note
object_info also includes the agent_info.
TaoWriter#
Overview#
TaoWriter overwrites IRABasicWriter’s bounding box output by applying a configurable occlusion check.
Parameters#
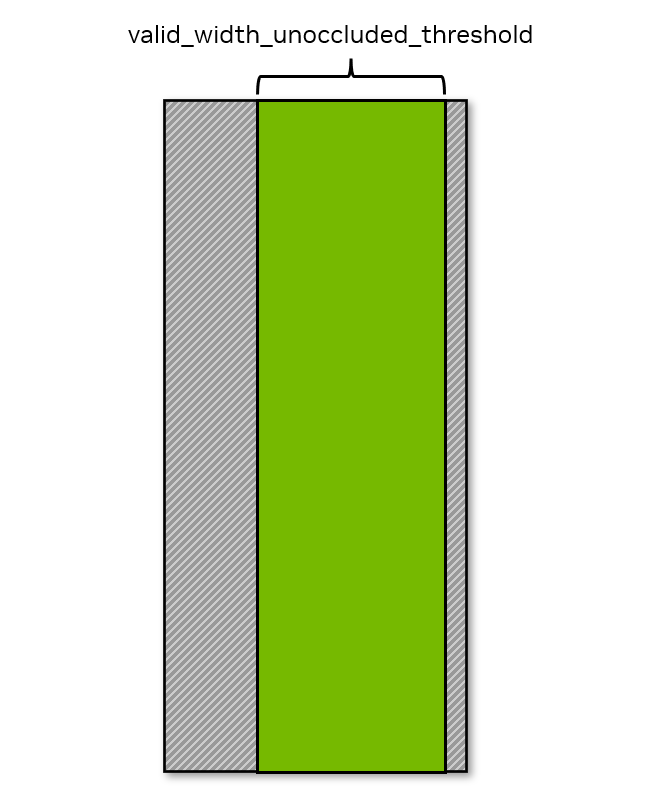
bbox: outputs objects and agents bounding box 2d and 3d data based on the occlusion rule mentioned below. It overwritesobject_info_bounding_box_2d_tight,object_info_bounding_box_2d_looseandobject_info_bounding_box_3dfromIRABasicWriter.shoulder_height_ratio: the ratio of head-to-shoulder distance to overall height.valid_width_unoccluded_threshold: the threshold for the ratio of visible character width to total width.valid_height_unoccluded_threshold: the threshold for the ratio of visible character height to total height.agent_info_skeleton_data: inherited from IRABasicWriter.
Note
shoulder_height_ratio, valid_width_unoccluded_threshold, and valid_height_unoccluded_threshold will be used to calculate the occlusion by the rule below. Occluded bounding box data will not be written to the output.
Occlusion Rule#
To make sure characters are mostly visible, the following test is applied to select valid characters for data generation.
Two conditions are defined for whether a character must be labeled, visibility in height and visibility in width. See the images for more details.
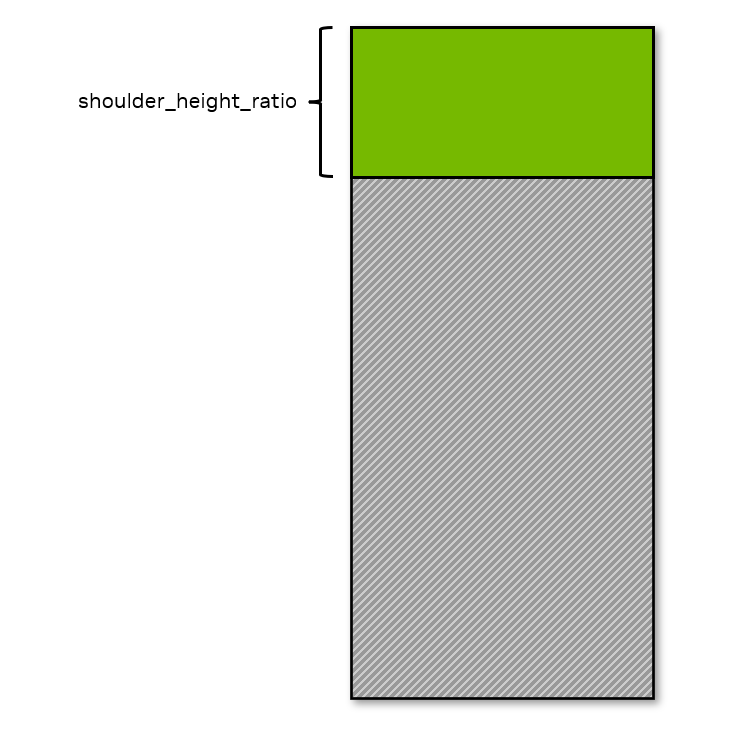
Visibility Requirement in Height
If more than
shoulder_height_ratioof the character’s upper body is visible then this condition is met.
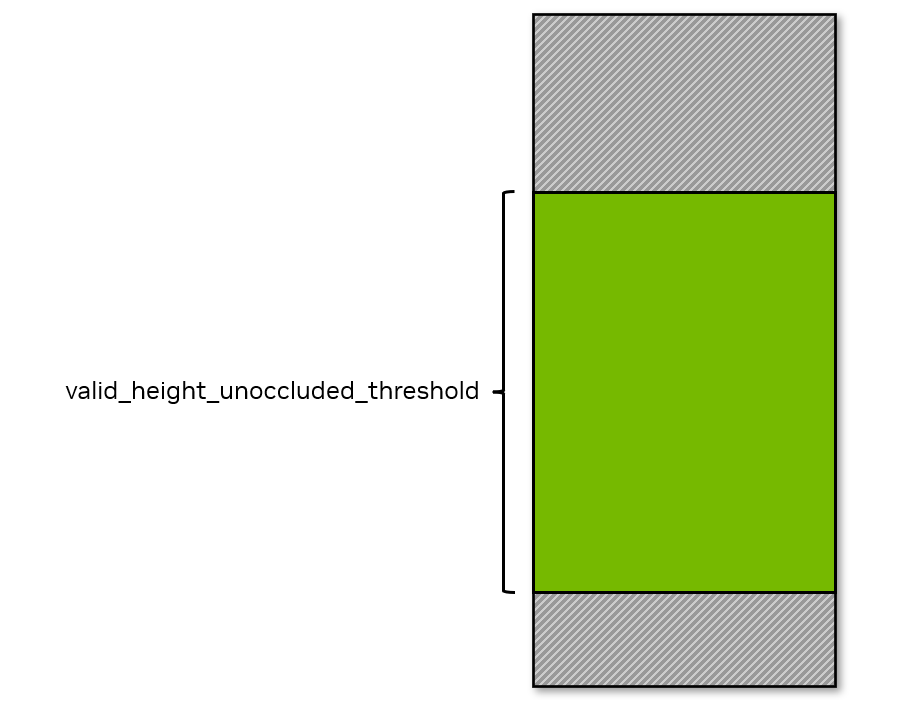
If more than
valid_height_unoccluded_thresholdof the character’s body height is visible, then this condition is met.
Note
Only one of the above conditions is needed to satisfy the height requirement.
Visibility Requirement in Width
Note
For occluded characters (characters that are blocked by an object within the camera frame), characters must satisfy both the visibility in height and width requirements.
For truncated characters (characters that are cut off by the camera frame), characters must satisfy visibility in height or the visibility in width requirement.
For characters that are both occluded and truncated, characters must satisfy the visibility in height or the visibility in width requirements.
StereoWriter#
Overview#
StereoWriter writes the data for simulating stereo cameras to the camera_params.json, while applying the same occlusion check as the TaoWriter.
Parameters#
customized_camera_params: output additional camera parameter data compared to the standardcamera_paramsannotator, such as the camera’s intrinsic data (fx_fy_cx_cy,focallength) and the stereo baseline (stereo_baseline).customized_distance_to_image_plane: output thedistance_to image_planedata in.pfmfile format.depth_format: specifies the data format fordistance_to_image_planeas either.pngor.npmifcustomized_distance_to_image_planeis not activated.bbox,shoulder_height_ratio,valid_height_unoccluded_threshold,valid_width_unoccluded_threshold, andagent_info_skeleton_dataare the same as the TaoWriter.
Note
Stereo cameras must be pairs with names like
Camera_01pairing withCamera_01_R, whereCamera_01is the left camera andCamera_01_Ris the right camera.IRA does not spawn stereo cameras. To create stereo cameras, you can run the following script.
DISTis the distance between the paired cameras, andCAMERA_PATHis the path to the camera that you want to make into a stereo camera. The script will treat the input camera as the left camera and generate the right camera for it.
DIST = 1
CAMERA_PATH = "/World/Cameras/Camera_01"
import omni.usd
import omni.kit.commands
from pxr import Gf, UsdGeom
camera_prim = omni.usd.get_context().get_stage().GetPrimAtPath(CAMERA_PATH)
new_translate = UsdGeom.Camera(camera_prim).GetCamera().transform.Transform(Gf.Vec3d(DIST,0,0))
omni.kit.commands.execute("CopyPrim", path_from=CAMERA_PATH, path_to=CAMERA_PATH+"_R")
stage.GetPrimAtPath(CAMERA_PATH + "_R").GetAttribute("xformOp:translate").Set(new_translate)
Custom Writers#
IRA supports using custom writers created by you, through the UI or config file directly.
To enable it from UI, select Custom from the dropdown of the Replicator Setting panel, enter the writer’s name and its input parameters in the text boxes.
To enable it from config file, put the writer’s name and parameters in the
replicatorsection.